Georgetown Researchers Uncover Key Differences in Bacterial Protein Regulation
Posted in News Story
A new study led by Dr. Rodrigo Maillard at Georgetown University, “Identifying Allosteric Hotspots in Mycobacterium tuberculosis cAMP Receptor Protein through Structural Homology,” published in Biochemistry sheds light on how bacteria regulate their genes, challenging long-held assumptions about protein behavior. The research compares how two bacterial species—Escherichia coli and Mycobacterium tuberculosis—use a signaling molecule called cyclic AMP (cAMP) to control important cellular functions.
Scientists have long believed that proteins with similar structures would function in the same way. However, this study found that even though the cAMP receptor proteins (CRPs) in E. coli and M. tuberculosis look alike, they behave very differently. By introducing targeted mutations into these proteins and studying their effects, the researchers discovered that each species has a unique way of responding to molecular signals.
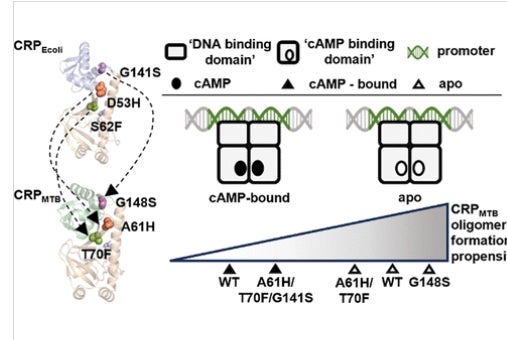
This study focuses on the differential allosteric regulation by the ubiquitous signaling molecule, cAMP, in the cAMP receptor protein from Escherichia coli (CRPEcoli) and from Mycobacterium tuberculosis (CRPMTB). By introducing structurally homologous mutations from allosteric hotspots previously identified in CRPEcoli into CRPMTB and examining their effects on protein solution structure, stability, and function, we aimed to determine the factors contributing to their differential allosteric regulation. Results from this study indicate that the structural similarity between two allosteric proteins from distantly related bacteria does not reliably predict their allosteric behavior nor identify allosteric hotspots involved in the response to molecular signals.
Why It Matters
Understanding how bacteria regulate their genes is crucial for many areas of science and medicine. M. tuberculosis, the bacterium that causes tuberculosis, relies on CRP for survival. By identifying differences in how these proteins function, scientists may be able to develop new drugs that specifically target M. tuberculosis without affecting other bacteria.
Looking Ahead
The findings open the door to further research on bacterial gene regulation and could lead to new ways to disrupt harmful bacterial processes. Future studies will explore how evolutionary differences shape these signaling pathways and whether similar variations exist in other bacteria
About the Researcher
Dr. Rodrigo Maillard is an Associate Professor in Georgetown’s Department of Chemistry and a member of the Institute for Soft Matter Synthesis and Metrology (ISMSM). His research focuses on understanding how proteins interact and function at the molecular level, using advanced techniques to study their behavior. His work has broad implications for drug discovery and bacterial gene regulation.
Read more HERE.