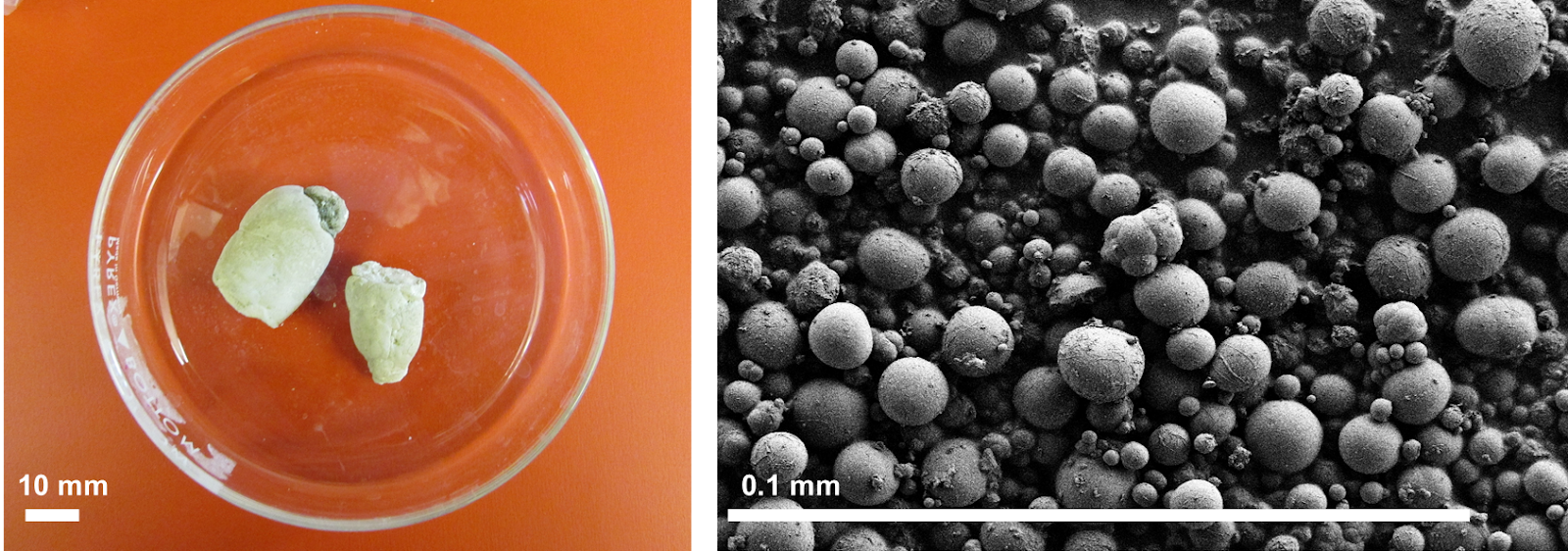
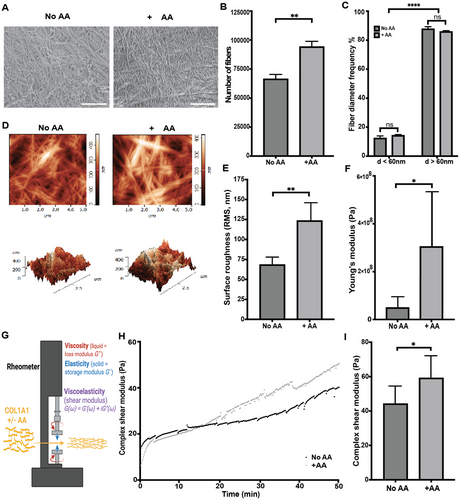
Welcoming Dr. Priyanka Joshi to the Institute for Soft Matter Synthesis and Metrology
February 11th, 2026
ISM is pleased to welcome Dr. Priyanka Joshi as a new Institute member. Dr. Joshi is a tenure-track Assistant Professor in the Department of Biochemistry and Molecular & Cellular Biology at Georgetown University Medical Center, where she directs the Laboratory of Biomolecular Homeostasis and Resilience.…